Welcome to See5, a system that extracts informative patterns from data. The following sections show how to prepare data files for See5 and illustrate the options for using the system.
We will illustrate See5 using a medical application -- mining a database of thyroid assays from the Garvan Institute of Medical Research, Sydney, to construct diagnostic rules for hypothyroidism. Each case concerns a single referral and contains information on the source of the referral, assays requested, patient data, and referring physician's comments. Here are three examples:
Attribute Case 1 Case 2 Case 3 .....
age 41 23 46
sex F F M
on thyroxine f f f
query on thyroxine f f f
on antithyroid medication f f f
sick f f f
pregnant f f not applicable
thyroid surgery f f f
I131 treatment f f f
query hypothyroid f f f
query hyperthyroid f f f
lithium f f f
tumor f f f
goitre f f f
hypopituitary f f f
psych f f f
TSH 1.3 4.1 0.98
T3 2.5 2 unknown
TT4 125 102 109
T4U 1.14 unknown 0.91
FTI 109 unknown unknown
referral source SVHC other other
diagnosis negative negative negative
ID 3733 1442 2965
This is exactly the sort of task for which See5 was designed. Each case belongs to one of a small number of mutually exclusive classes (negative, primary, secondary, compensated). Properties of every case that may be relevant to its class are provided, although some cases may have unknown or non-applicable values for some attributes. There are 24 attributes in this example, but See5 can deal with any number of attributes.
See5's job is to find how to predict a case's class from the values of the other attributes. See5 does this by constructing a classifier that makes this prediction. As we will see, See5 can construct classifiers expressed as decision trees or as sets of rules.
hypothyroid
for this illustration.
All files read or written by See5 for an application
have names of the form filestem.extension,
where filestem identifies the application and
extension describes the contents of the file.
The case of letters in both the filestem and extension is important --
file names APP.DATA,
app.data, and App.Data, are all different.
It is important that the extensions are written
exactly as shown below, otherwise See5 will not recognize
the files for your application.
hypothyroid.names) that
describes the attributes and classes.
There are two important subgroups of attributes:
The file hypothyroid.names looks like this:
diagnosis. | the target attribute
age: continuous.
sex: M, F.
on thyroxine: f, t.
query on thyroxine: f, t.
on antithyroid medication: f, t.
sick: f, t.
pregnant: f, t.
thyroid surgery: f, t.
I131 treatment: f, t.
query hypothyroid: f, t.
query hyperthyroid: f, t.
lithium: f, t.
tumor: f, t.
goitre: f, t.
hypopituitary: f, t.
psych: f, t.
TSH: continuous.
T3: continuous.
TT4: continuous.
T4U: continuous.
FTI:= TT4 / T4U.
referral source: WEST, STMW, SVHC, SVI, SVHD, other.
diagnosis: primary, compensated, secondary, negative.
ID: label.
|')
can appear
in names and values, but must be prefixed by the escape character
`\'.
For example, the name "Filch, Grabbit, and Co." would be written
as Filch\, Grabbit\, and Co\.'|'
causes the remainder of the line to be ignored and is handy for
including comments.
This use of `|' should not occur inside a value.
The first line of the names file gives the classes, either by naming a discrete attribute (the target attribute) that contains the class value (as in this example), or by listing them explicitly. The attributes are then defined in the order that they will be given for each case.
The name of each explicitly-defined attribute is followed by a colon `:' and a description of the values taken by the attribute. There are six possibilities:
continuous
date
YYYY/MM/DD
or YYYY-MM-DD,
e.g. 1999/09/30 or 1999-09-30.
Valid dates range from the year 1601
to the year 4000 (perhaps optimistically assuming that
humanity survives as long as that!).
[ordered] to indicate
that they are given in a meaningful ordering, otherwise they will
be taken as unordered. For instance, the values low, medium, high
are ordered, while
meat, poultry, fish, vegetables are not.
The former might be declared as
grade: [ordered] low, medium, high.
discrete N for some integer N
ignore
label
The name of each implicitly-defined attribute is followed by `:=' and then a formula defining the attribute value. The formula is written in the usual way, using parentheses where needed, and may refer to any attribute defined before this one. Constants in the formula can be numbers (written in decimal notation), dates, and discrete attribute values (enclosed in string quotes `"'). The operators and functions that can be used in the formula are
FTI
above is continuous, since its value is obtained by dividing one number by
another. The value of a hypothetical attribute such as
strange := referral source = "WEST" or age > 40.
t or f
since the value given by the formula is either true or false.
Dates are stored by See5 as the number of days since a particular
starting point (1600/03/01)
so some operations on dates make sense.
Thus, if we have attributes
d1: date.
d2: date.
interval := d2 - d1.
gap := d1 <= d2 - 7.
d1-day-of-week := (d1 + 1) % 7 + 1.
interval then represents the number of days from
d1 to d2 (non-inclusive) and
gap would have a true/false value signaling whether
d1 is at least a week before d2.
The last definition is a slightly non-obvious way of determining
the day of the week on which d1 falls, with values
ranging from 1 (Monday) to 7 (Sunday).
Finally, if the value of the formula cannot be determined for a particular case because one or more of the attributes appearing in the formula have unknown values, the value of the implicitly-defined attribute is also unknown.
hypothyroid.data)
provides information on the
training
cases from which See5 will extract patterns.
The entry for each case consists of one or more lines that give
the values for all explicitly-defined attributes. If the classes are listed
in the first line of the names file,
the attribute values are followed by the case's class value.
Values are separated by commas and the entry is optionally terminated by
a period.
Once again, anything on a line after a vertical bar `|'
is ignored.
(If the information for a case occupies more than one line, make sure
that the line breaks occur after commas.)
For example,
the first three cases from file hypothyroid.data are:
41,F,f,f,f,f,f,f,f,f,f,f,f,f,f,f,1.3,2.5,125,1.14,SVHC,negative,3733
23,F,f,f,f,f,f,f,f,f,f,f,f,f,f,f,4.1,2,102,?,other,negative,1442
46,M,f,f,f,f,N/A,f,f,f,f,f,f,f,f,f,0.98,?,109,0.91,other,negative,2965
Notice that
`?' is used to denote a value that is missing or unknown.
Similarly, `N/A' denotes a value that is not applicable for
a particular case.
Also note
that the cases do not contain values for the attribute FTI
since its values are computed from other attribute values.
The third kind of file used
by See5 consists of new test
cases (e.g. hypothyroid.test) on which the classifier
can be evaluated.
This file is optional and, if used, has
exactly the same format as the data file.
Another optional file, the cases file
(e.g. hypothyroid.cases),
differs from a test file only in allowing the cases'
classes to be unknown (`?').
The cases file is used primarily with
the cross-referencing procedure and public source code,
both of which are
described later on.
hypothyroid.costs),
is also optional and sets out
differential misclassification costs.
In some applications
there is a much higher penalty for certain types of mistakes.
In this application, a prediction that hypothyroidism is not present
could be very costly if in fact it is.
On the other hand, predicting incorrectly that a patient is
hypothyroid
may be a less serious error.
See5 allows different misclassification
costs to be associated with each combination of real class and
predicted class. We will return to this topic near the end of the
tutorial.
It is difficult to see what is going on in an interface without actually using it. As a simple illustration, here is the main window of See5 after the hypothyroid application has been selected.

The main window of See5 has six buttons on its toolbar. From left to right, they are
The Edit menu facilities changes to the names and costs files after an application's files have been located. On-line help is available through the Help menu.
The first step is to locate the date using the Locate Data button on the toolbar (or the corresponding selection from the File menu). We will assume that the hypothyroid data above has been located in this manner.
There are several options that affect the type of classifier that See5 produces and the way that it is constructed. The Construct Classifier button on the toolbar (or selection from the File menu) displays a dialog box that sets out these classifier construction options:
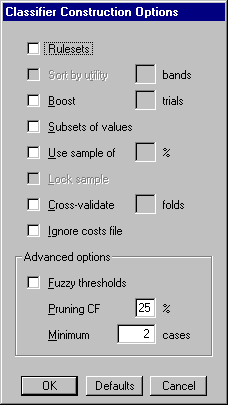
In this section we will examine them in turn, starting with the simpler situations.
See5 [Release 1.12] Thu Feb 17 15:13:04 2000
-------------------
Class specified by attribute `diagnosis'
Read 2772 cases (24 attributes) from hypothyroid.data
Decision tree:
TSH <= 6: negative (2472/2)
TSH > 6:
:...FTI > 65:
:...on thyroxine = t: negative (37.7)
: on thyroxine = f:
: :...thyroid surgery = t: negative (6.8)
: thyroid surgery = f:
: :...TT4 > 153: negative (6/0.1)
: TT4 <= 153:
: :...TT4 <= 37: primary (2.5/0.2)
: TT4 > 37: compensated (174.6/24.8)
FTI <= 65:
:...thyroid surgery = t:
:...TT4 <= 48: negative (2)
: TT4 > 48: primary (2.2/0.2)
thyroid surgery = f:
:...TT4 <= 61: primary (51/3.7)
TT4 > 61:
:...referral source in {WEST,SVHD}: primary (0)
referral source = STMW: primary (0.1)
referral source = SVHC: primary (1)
referral source = SVI: primary (3.8/0.8)
referral source = other:
:...TSH > 22: primary (5.8/0.8)
TSH <= 22:
:...T3 <= 2.3: compensated (3.4/0.9)
T3 > 2.3: negative (3/0.2)
Evaluation on training data (2772 cases):
Decision Tree
----------------
Size Errors
16 6( 0.2%) <<
(a) (b) (c) (d) <-classified as
---- ---- ---- ----
60 3 (a): class primary
154 (b): class compensated
2 (c): class secondary
1 2552 (d): class negative
Evaluation on test data (1000 cases):
Decision Tree
----------------
Size Errors
16 4( 0.4%) <<
(a) (b) (c) (d) <-classified as
---- ---- ---- ----
31 1 (a): class primary
1 39 (b): class compensated
(c): class secondary
2 926 (d): class negative
Time: 0.3 secs
The first line identifies the version of See5 and the run date.
See5 constructs a decision tree from the 2772 training cases
in the file hypothyroid.data, and this appears next.
Although it may not look much like a tree, this output can be
paraphrased as:
if TSH is less than or equal to 6 then negative else if TSH is greater than 6 then if FTI is greater than 65 then if on thyroxine equals t then negative else if on thyroxine equals f then if thyroid surgery equals t then negative else if thyroid surgery equals f then if TT4 is greater than 153 then negative else if TT4 is less than or equal to 153 then if TT4 is less than or equal to 37 then primary else if TT4 is greater than 37 then compensated else if FTI is less than or equal to 65 then . . . .and so on. The tree employs a case's attribute values to map it to a leaf designating one of the classes. Every leaf of the tree is followed by a cryptic (n) or (n/m). For instance, the last leaf of the decision tree is
negative (3/0.2), for which n is 3 and
m is 0.2.
The value of n is the number of cases in the file
hypothyroid.data
that are mapped to this leaf, and m (if it appears) is the number of
them that are classified incorrectly by the leaf.
(A non-integral number of cases can arise because, when the value of
an attribute in the tree is not known, See5 splits the case
and sends a fraction down each branch.)
The last section of the See5 output concerns the evaluation of the
decision tree, first on the cases in hypothyroid.data from which
it was constructed, and then on the new cases in hypothyroid.test.
The size of the tree is its number of leaves and the
column headed Errors shows
the number and percentage of cases misclassified.
The tree, with 16 leaves, misclassifies 6 of the 2772 given cases, an error
rate of 0.2%.
(This might seem inconsistent with the errors recorded at the leaves --
one of the leaves shows 24.8 errors! The discrepancy arises because
parts of cases split as above can be misclassified and yet, when the
votes from all the parts are aggregated, the correct class can still be
chosen.)
Performance on the training cases is further analyzed in a
confusion matrix
that pinpoints the kinds of errors made.
In this example, the decision tree misclassifies
three of the primary cases as compensated,
both secondary cases as negative,
and one negative case as compensated.
A very simple majority classifier predicts
that every new case belongs to the most common class in the
training data.
In this example, 2553 of the 2772 training cases belong to class
negative so that
a majority classifier would always opt for negative.
The 1000 test cases from file hypothyroid.test
include 928 belonging to class negative, so a simple
majority classifier would have an error rate of 7.2%.
The decision tree has a lower error rate of 0.4% on the new
cases, but notice that
this is higher than its error rate on the training cases.
The confusion matrix for the test cases again shows
the detailed breakdown of correct and incorrect classifications.
The default way in which See5 constructs tests on discrete attributes as to associate a separate branch with each value for which cases are available. Tests with a high fan-out can have the undesirable side-effect of fragmenting the data during construction of the decision tree. See5 has a Subset option that can mitigate this fragmentation to some extent: attribute values are grouped into subsets and each subtree is associated with a subset rather than with a single value.
In the hypothyroid example, invoking this option has little effect on the tree, merely simplifying the later part to
referral source in {WEST,STMW,SVHC,SVI,SVHD}: primary (4.9/0.8)
referral source = other:
:...TSH > 22: primary (5.8/0.8)
TSH <= 22:
:...T3 <= 2.3: compensated (3.4/0.9)
T3 > 2.3: negative (3/0.2)
Decision trees can sometimes be very difficult to understand. An important feature of See5 is its mechanism to convert trees into collections of rules called rulesets. The Rulesets option causes rules to be derived from trees produced as above, giving the following rules:
Rule 1: (31, lift 42.7)
thyroid surgery = f
TSH > 6
TT4 <= 37
-> class primary [0.970]
Rule 2: (63/6, lift 39.3)
TSH > 6
FTI <= 65
-> class primary [0.892]
Rule 3: (141/1, lift 17.7)
on thyroxine = f
thyroid surgery = f
TSH > 6
TT4 <= 153
FTI > 65
-> class compensated [0.986]
Rule 4: (114/24, lift 14.1)
thyroid surgery = f
TSH > 6
TSH <= 22
T3 <= 2.3
TT4 > 61
-> class compensated [0.784]
Rule 5: (2225/2, lift 1.1)
TSH <= 6
-> class negative [0.999]
Rule 6: (296, lift 1.1)
on thyroxine = t
FTI > 65
-> class negative [0.997]
Rule 7: (240, lift 1.1)
TT4 > 153
-> class negative [0.996]
Rule 8: (531/16, lift 1.1)
TSH <= 22
T3 > 2.3
TT4 > 61
-> class negative [0.968]
Rule 9: (38/2, lift 1.0)
thyroid surgery = t
-> class negative [0.925]
Default class: negative
Each rule consists of:
negative,
that is used when none of the rules apply.
Rulesets are generally much simpler to understand than trees since each rule describes a specific context associated with a class. Furthermore, a ruleset generated from a tree usually has fewer rules than than the tree has leaves, another plus for comprehensibility. (In this example, the first decision tree with 16 leaves is reduced to nine rules.) Finally, rules are often more accurate predictors than decision trees -- a point not illustrated here, since both have an error rate of 0.4% on the test cases. For very large datasets, however, generating rules with the Ruleset option can require considerably more computer time.
In the example above, rules are ordered by class and
sub-ordered by confidence.
This is the default, but an alternative ordering by
contribution to predictive accuracy
can be selected using the
Sort by utility
option. Under this option, the rule that most reduces the error rate
appears first and the rule that contributes least appears last.
Furthermore, results are reported in a selected number of
bands so that the predictive accuracies of the more
important subsets of rules are also estimated. For example,
if the
Sort by utility option with 5 bands
is selected, the hypothyroid rules are reordered as
Rule 1: (2225/2, lift 1.1)
TSH <= 6
-> class negative [0.999]
Rule 2: (141/1, lift 17.7)
on thyroxine = f
thyroid surgery = f
TSH > 6
TT4 <= 153
FTI > 65
-> class compensated [0.986]
Rule 3: (63/6, lift 39.3)
TSH > 6
FTI <= 65
-> class primary [0.892]
Rule 4: (296, lift 1.1)
on thyroxine = t
FTI > 65
-> class negative [0.997]
Rule 5: (240, lift 1.1)
TT4 > 153
-> class negative [0.996]
Rule 6: (114/24, lift 14.1)
thyroid surgery = f
TSH > 6
TSH <= 22
T3 <= 2.3
TT4 > 61
-> class compensated [0.784]
Rule 7: (38/2, lift 1.0)
thyroid surgery = t
-> class negative [0.925]
Rule 8: (31, lift 42.7)
thyroid surgery = f
TSH > 6
TT4 <= 37
-> class primary [0.970]
Rule 9: (531/16, lift 1.1)
TSH <= 22
T3 > 2.3
TT4 > 61
-> class negative [0.968]
Rule utility summary:
Rules Errors
----- ------
1-2 39( 3.9%)
1-4 9( 0.9%)
1-5 9( 0.9%)
1-7 5( 0.5%)
Another innovation incorporated in See5 is adaptive boosting, based on the work of Rob Schapire and Yoav Freund. The idea is to generate several classifiers (either decision trees or rulesets) rather than just one. When a new case is to be classified, each classifier votes for its predicted class and the votes are counted to determine the final class.
But how can we generate several classifiers from a single dataset?
As the first step, a single decision tree or ruleset is constructed
as before from the training data (e.g. hypothyroid.data).
This classifier will usually make mistakes on some cases in the data;
the first decision tree, for instance, gives the wrong class
for 6 cases in hypothyroid.data.
When the second classifier is constructed, more attention is paid
to these cases in an attempt to get them right.
As a consequence, the second classifier will generally be different
from the first. It also will make errors on some cases,
and these become the the focus of attention during construction
of the third classifier.
This process continues for a pre-determined number of iterations.
The Boost option with x trials instructs See5 to construct up to x classifiers in this manner. Naturally, constructing multiple classifiers requires more computation that building a single classifier -- but the effort can pay dividends! Trials over numerous datasets, large and small, show that on average 10-classifier boosting reduces the error rate for test cases by about 25%.
Selecting the Rulesets option and the Boost option with 10 trials
causes ten rulesets to be generated. The summary of the rulesets'
individual and aggregated performance on the 1000 test cases is:
Trial Decision Tree Rules
----- ---------------- ----------------
Size Errors No Errors
0 16 4( 0.4%) 9 4( 0.4%)
1 27 31( 3.1%) 19 20( 2.0%)
2 44 24( 2.4%) 20 4( 0.4%)
3 31 31( 3.1%) 19 19( 1.9%)
4 35 16( 1.6%) 19 7( 0.7%)
5 31 25( 2.5%) 20 12( 1.2%)
6 38 13( 1.3%) 23 11( 1.1%)
7 37 12( 1.2%) 20 7( 0.7%)
8 39 13( 1.3%) 21 7( 0.7%)
9 32 30( 3.0%) 17 20( 2.0%)
boost 7( 0.7%) 3( 0.3%) <<
(Again, different floating point hardware can lead to slightly different
results.)
The performance of the classifier constructed at each iteration or trial
is summarized on a separate line, while the line labeled
boost
shows the result of voting all the classifiers.
The tree and ruleset constructed on Trial 0 are identical to those
produced without the Boost option.
Some of the subsequent trees and rulesets produced by paying more attention
to certain cases
have quite high overall error rates. When the
ten rulesets are combined by voting, however,
the final predictions have an error rate of 0.3% on
the test cases.
The top of our initial decision tree tests whether
the value of the attribute TSH is less than or
equal to, or greater than, 6. If the former holds, we go no further
and predict that the case's class is negative, while
if it does not we look at other information before making a decision.
Thresholds like this are sharp by default, so that a case with
a hypothetical value of 5.99 for TSH is treated
quite differently from one with a value of 6.01.
For some domains, this sudden change is quite appropriate -- for instance, there are hard-and-fast cutoffs for bands of the income tax table. For other applications, though, it is more reasonable to expect classification decisions to change more slowly with changes in attribute values.
See5 contains an option to `soften' thresholds such as 6 above. When this is invoked, each threshold is broken into three ranges -- let us denote them by a lower bound lb, an upper bound ub, and a central value t. If the attribute value in question is below lb or above ub, classification is carried out using the single branch corresponding to the `<=' or '>' result respectively. If the value lies between lb and ub, both branches of the tree are investigated and the results combined probabilistically. The values of lb and ub are determined by See5 based on an analysis of the apparent sensitivity of classification to small changes in the threshold. They need not be symmetric -- a fuzzy threshold can be relatively hard in one direction.
Invoking the Fuzzy thresholds option
gives the following decision tree:
TSH <= 6 (6.05): negative (2472/2)
TSH >= 6.1 (6.05):
:...FTI >= 65.7 (65.35):
:...on thyroxine = t: negative (37.7)
: on thyroxine = f:
: :...thyroid surgery = t: negative (6.8)
: thyroid surgery = f:
: :...TT4 >= 158 (153): negative (6/0.1)
: TT4 <= 148 (153):
: :...TT4 <= 31 (37.5): primary (2.5/0.2)
: TT4 >= 44 (37.5): compensated (174.6/24.8)
FTI <= 65 (65.35):
:...thyroid surgery = t:
:...TT4 <= 36 (49.5): negative (2)
: TT4 >= 63 (49.5): primary (2.2/0.2)
thyroid surgery = f:
:...TT4 <= 60 (61.5): primary (51/3.7)
TT4 >= 63 (61.5):
:...referral source in {WEST,SVHD}: primary (0)
referral source = STMW: primary (0.1)
referral source = SVHC: primary (1)
referral source = SVI: primary (3.8/0.8)
referral source = other:
:...TSH >= 44 (22.5): primary (5.8/0.8)
TSH <= 19 (22.5):
:...T3 <= 2.3 (2.35): compensated (3.4/0.9)
T3 >= 2.4 (2.35): negative (3/0.2)
<= lb (t)
or >= ub (t). In this example, most
of the thresholds are still relatively tight, but notice
the asymmetric threshold values for the last test of TSH.
Soft thresholds do not affect accuracy on this test set.
A final point: soft thresholds affect only decision tree classifiers -- they do not change the interpretation of rulesets.
Two further options enable aspects of the classifier-generation process to be tweaked. These are best regarded as advanced options that should be used sparingly (if at all), so that this section can be skipped without much loss.
See5 constructs decision trees in two phases. A large tree is first grown to fit the data closely and is then `pruned' by removing parts that are predicted to have a relatively high error rate. The Pruning CF option affects the way that error rates are estimated and hence the severity of pruning; values smaller than the default (25%) cause more of the initial tree to be pruned, while larger values result in less pruning.
The Minimum cases option constrains the degree to which the initial tree can fit the data. At each branch point in the decision tree, the stated minimum number of training cases must follow at least two of the branches. Values higher than the default (2 cases) can lead to an initial tree that fits the training data only approximately -- a form of pre-pruning. (This option is complicated by the presence of missing attribute values and by the use of differential misclassification costs, discussed below. Both cause adjustments to the apparent number of cases following a branch.)
Even though See5 is relatively fast, building classifiers from large numbers of cases can take an inconveniently long time, especially when options such as boosting are employed. See5 incorporates a facility to extract a random sample from a dataset, construct a classifier from the sample, and then test the classifier on a disjoint collection of cases. By using a smaller set of training cases in this way, the process of generating a classifier is expedited, but at the cost of a possible reduction in the classifier's predictive performance.
The Sample option with x% has two consequences. Firstly, a random sample containing x% of the cases in the application's data file is used to construct the classifier. Secondly, the classifier is evaluated on a non-overlapping set of test cases consisting of another (disjoint) sample of the same size as the training set (if x is less than 50%), or all cases that were not used in the training set (if x is greater than or equal to 50%).
In the hypothyroid example,
using a sample of 60% would cause a classifier to be constructed
from a randomly-selected 1663 of the 2772 cases in
hypothyroid.data, then tested on the
remaining 1109 cases.
By default, the random sample changes every time that a classifier is constructed, so that successive runs of See5 with sampling will usually produce different results. This resampling can be avoided by selecting the Lock sample option that uses the current sample for constructing subsequent classifiers. If this option is selected, the sample will change only when another application is loaded, the sample percentage is altered, the option is unselected, or See5 is restarted.
As we saw earlier, the performance of a classifier on the training
cases from which it was constructed gives a poor estimate of
its accuracy on new cases.
The true predictive accuracy of the classifier can be estimated
by sampling, as above, or by using a separate test file;
either way, the classifier is evaluated on cases that were
not used to build it.
However, this estimate can be unreliable unless the numbers of
cases used to build and evaluate the classifier are both large.
If the cases in hypothyroid.data and
hypothyroid.test were to be shuffled
and divided into a new 2772-case training set and a 1000-case test set,
See5 might construct a different classifier with a lower or higher error
rate on the test cases.
One way to get a more reliable estimate of predictive accuracy is by f-fold cross-validation. The cases in the data file are divided into f blocks of roughly the same size and class distribution. For each block in turn, a classifier is constructed from the cases in the remaining blocks and tested on the cases in the hold-out block. In this way, each case is used just once as a test case. The error rate of a classifier produced from all the cases is estimated as the ratio of the total number of errors on the hold-out cases to the total number of cases.
The Cross-validation option with f folds runs such a f-fold cross-validation. Suppose now that we select the Cross-validation option with 10 folds together with the Rulesets option. After giving details of the individual decision trees and rulesets, the output shows a summary like this:
Fold Decision Tree Rules
---- ---------------- ----------------
Size Errors No Errors
0 10 0.3% 7 0.3%
1 8 1.1% 7 1.1%
2 12 0.3% 7 0.3%
3 9 0.0% 7 0.0%
4 13 0.3% 7 0.3%
5 9 0.3% 7 0.3%
6 9 0.5% 7 0.8%
7 10 0.5% 7 0.8%
8 9 1.1% 8 1.1%
9 10 0.5% 7 0.8%
Mean 9.9 0.5% 7.1 0.6%
SE 0.5 0.1% 0.1 0.1%
This estimates the error rate of decision tree and ruleset classifiers
produced from the
2772 cases in hypothyroid.data
at 0.5% and 0.6% respectively.
The SE figures (the standard errors of the means)
provide an estimate of the variability of these results.
The cross-validation procedure can be repeated for different random partitions of the cases into blocks. The average error rate from these distinct cross-validations is then an even more reliable estimate of the error rate of the single classifier produced from all the cases.
Since every cross-validation fold uses only part of the application's data, running a cross-validation does not result in a classifier being saved. To save a classifier for later use, simply run See5 without employing cross-validation.
Up to this point, all errors have been treated as equal -- we have simply counted the number of errors made by a classifier to summarize its performance. Let us now turn to the situation in which the `cost' associated with a classification error depends on the predicted and true class of the misclassified case.
See5 allows costs to be assigned to any combination of predicted and true class via entries in the optional file filestem.costs. Each entry has the form
predicted class, true class: cost
where cost is a non-negative real number. The file may contain any number of entries; if a particular combination is not specified explicitly, its cost is taken to be 0 if the predicted class is correct and 1 otherwise.
To illustrate the idea, suppose that it was a much more serious
error to classify a hypothyroid patient as negative
than the converse.
A hypothetical costs file hypothyroid.costs
might look like this:
negative, primary: 8
negative, secondary: 8
negative, compensated: 8
This specifies that the cost of misclassifying any
primary,
secondary, or
compensated
patient as negative is 8 units.
Since they are not given explicitly, all other errors
have cost 1 unit.
In other words, the first kind of error is 8 times more costly.
A costs file is automatically read by See5 unless the system is told to ignore it. The output from the system using default options now looks like this:
See5 [Release 1.12] Thu Feb 17 15:27:12 2000
-------------------
Class specified by attribute `diagnosis'
Read 2772 cases (24 attributes) from hypothyroid.data
Read misclassification costs from hypothyroid.costs
Decision tree:
TSH <= 6:
:...query hypothyroid = t:
: :...TT4 <= 34: secondary (1.1/0.1)
: : TT4 > 34: negative (122.3)
: query hypothyroid = f:
: :...TSH <= 4.5: negative (2267.3)
: TSH > 4.5:
: :...TT4 > 50: negative (78.2)
: TT4 <= 50:
: :...referral source in {WEST,STMW,SVHC,SVHD}: secondary (0)
: referral source = SVI: negative (2)
: referral source = other: secondary (1.2/0.2)
TSH > 6:
:...FTI <= 65:
:...TT4 <= 63: primary (59.4/8)
: TT4 > 63:
: :...TSH <= 9.4: compensated (3.3/1.3)
: TSH > 9.4:
: :...TT4 > 90: compensated (1.1/0.2)
: TT4 <= 90:
: :...pregnant = t: primary (0)
: pregnant = f: primary (7/1)
: pregnant = N/A:
: :...TT4 <= 75: compensated (0.5)
: TT4 > 75: primary (1.1/0.1)
FTI > 65:
:...on thyroxine = t: negative (37.7)
on thyroxine = f:
:...thyroid surgery = t: negative (6.8)
thyroid surgery = f:
:...TT4 <= 62:
:...TSH <= 35: compensated (4.5/0.4)
: TSH > 35: primary (2.5/0.2)
TT4 > 62:
:...age > 8:
:...TT4 <= 153: compensated (164.9/22.9)
: TT4 > 153: negative (6/0.1)
age <= 8:
:...TSH > 29: primary (0.7)
TSH <= 29:
:...referral source in {WEST,SVHC,SVI,
: SVHD}: compensated (0)
referral source = other: compensated (2.8)
referral source = STMW:
:...age <= 1: compensated (1)
age > 1: primary (0.7)
Evaluation on training data (2772 cases):
Decision Tree
-----------------------
Size Errors Cost
25 13( 0.5%) 0.00 <<
(a) (b) (c) (d) <-classified as
---- ---- ---- ----
62 1 (a): class primary
154 (b): class compensated
2 (c): class secondary
6 6 2541 (d): class negative
Evaluation on test data (1000 cases):
Decision Tree
-----------------------
Size Errors Cost
25 8( 0.8%) 0.01 <<
(a) (b) (c) (d) <-classified as
---- ---- ---- ----
32 (a): class primary
1 39 (b): class compensated
(c): class secondary
1 6 921 (d): class negative
Time: 0.5 secs
hypothyroid.data.
The corresponding value for the new tree is 13, since no
non-negative cases are classified as negative.
That is, the total misclassification cost over the
training cases is lower than that of the old tree.
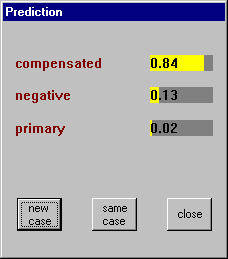
Once a classifier has been constructed, an interactive interpreter can be used to assign new cases to classes. The Use Classifier button invokes the interpreter, using the most recent classifier for the current application, and prompts for information about the case to be classified. Since the values of all attributes may not be needed, the attribute values requested will depend on the case itself. When all the relevant information has been entered, the most probable class (or classes) are shown, each with a certainty value. For example, this is the result of analyzing a case using the first decision tree above:
See5 incorporates a unique facility that links data and the
relevant sections of (possibly boosted) classifiers.
We will illustrate this facility using the first decision tree
for the hypothyroid application and the cases in hypothyroid.data
from which it was constructed.
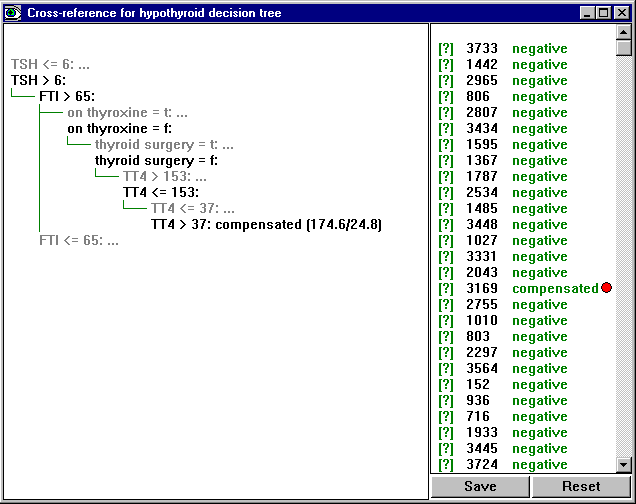
The Cross-Reference button brings up a window showing the most recent classifier for the current application and how it relates to the cases in the data, test or cases file. (If more than one of these is present, a menu will prompt you to select the file.)
The window is divided into two panes, with the classifier on the left and a list of cases on the right. Each case has a [?] tag (that is red if the case is misclassified), an identifying number or label, and the actual class of the case. Clicking on a case's label or number shows the part(s) of the classifier(s) relevant to that case. For instance, clicking on case 3169 shows the leaf to which this case is mapped:
If a case has missing values for one or more attributes, if it is covered by several rules, or if boosted classifiers are used, more than one leaf or rule may be relevant to a case. In such situations, all relevant classifier parts are shown.
Click on any leaf or rule, and all the cases that map to the leaf or rule are shown. For instance, clicking on the third leaf from the top shows all cases that are covered by the leaf:
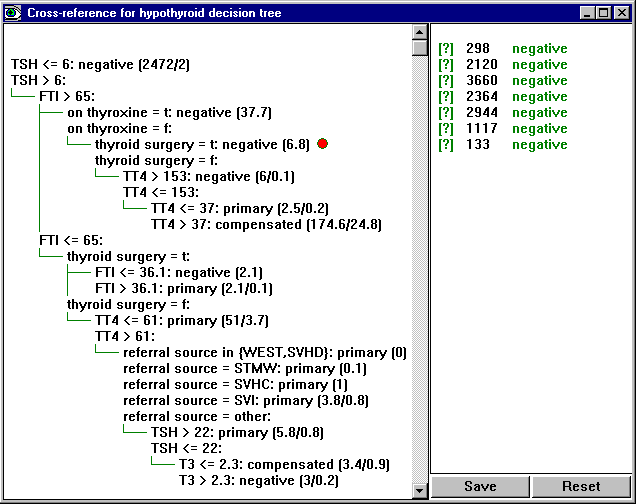
The Save button preserves the details of the displayed model and case list as an ascii file -- the file name is selected through a dialog box. The Reset button can be used to restore the window to its initial state. Finally, clicking on the tag [?] in front of a case number or label displays that case:
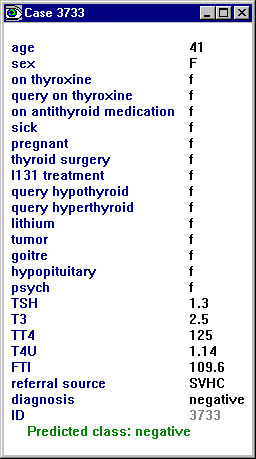
The values of attributes marked ignore or label
are displayed in a lighter tone to indicate that they play no part
in classifying the case.
The See5 distribution includes a program See5X that
can be used to produce models non-interactively.
This console application resides in the same folder as See5
(usually C:\Program Files\See5 for single-computer licences
or the See5 folder on your desktop for network licences)
and is invoked from
an MS-DOS Prompt window. The command to run the program is:
start See5X -f filestem parameters
where the parameters enable one or more options discussed above to
be selected:
-s use the Subset option -r use the Ruleset option -u sort rules by their utility -b use the Boosting option with 10 trials -t trials ditto with specified number of trials -S x use the Sampling option with x% -I seed set the sampling seed value -X folds carry out a cross-validation -c CF set the Pruning CF value -m cases set the Minimum cases -p use the Fuzzy thresholds option -e ignore any costs file -h print a summary of the batch mode options
If desired, output from See5 can be diverted to a file in the usual way.
As an example (for a single-computer licensee), typing the commands
cd "C:\Program Files\See5"
start See5X -f Samples\anneal -r -b >save.txt
save.txt.
The classifiers generated by See5 are retained in files filestem.tree (for decision trees) and filestem.rules (for rulesets). Free C source code is available to read these classifier files and to make predictions with them. Using this code, it is possible to call See5 classifiers from other programs. As an example, the source includes a program to read cases from a cases file, and to show how each is classified by boosted or single trees or rulesets.
Click here to download a WinZip archive containing the public source.
| © RULEQUEST RESEARCH 2000 | Last updated February 2000 |
| home | products | download | evaluations | prices | purchase | contact us |